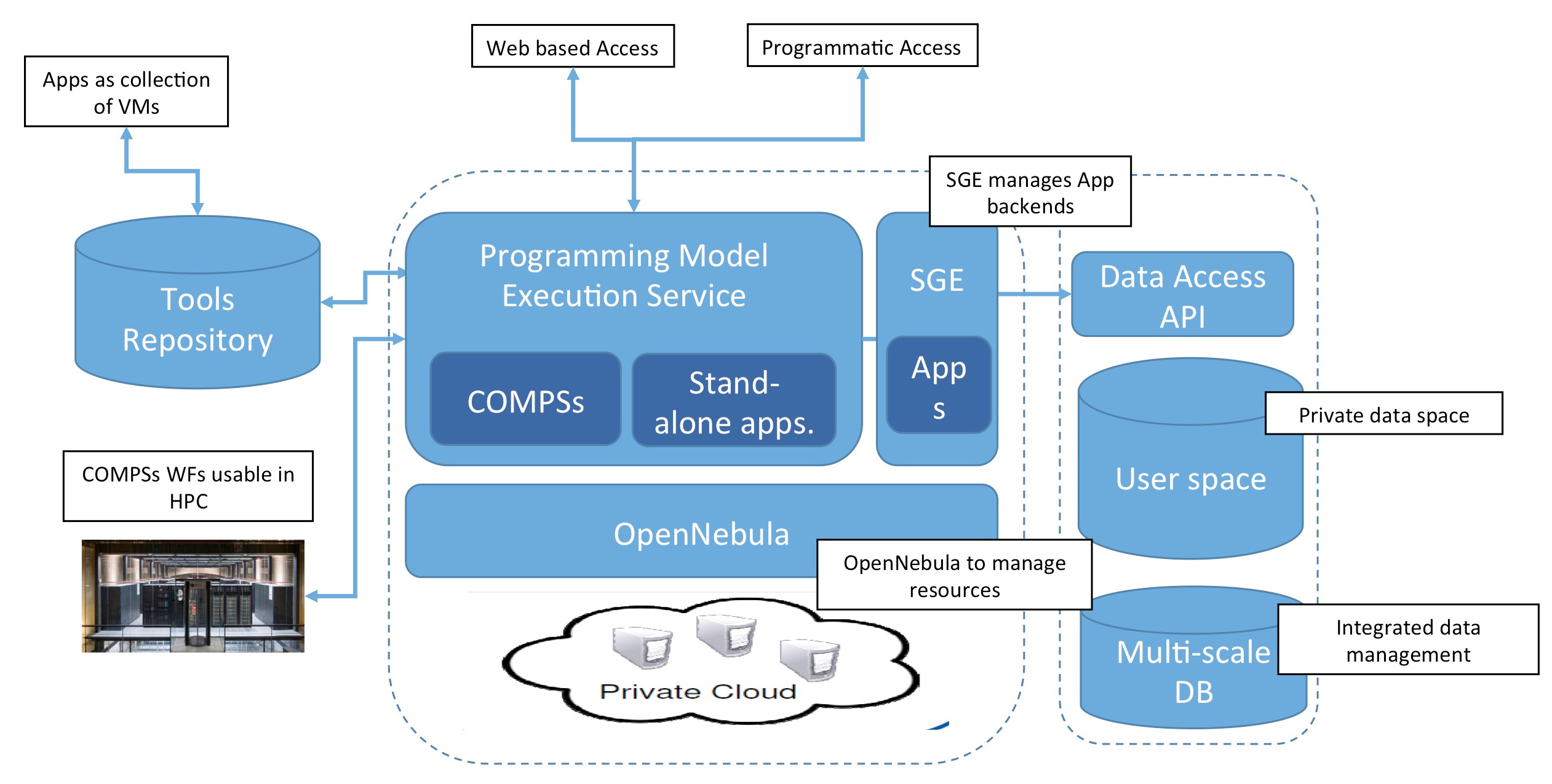
Workflows are defined by the use of COMPSs programming model. COMPSs is able to discover implicit parallelism in the pipelines, and hence, execute otherwise serial operations with an optimal use of a parallel environment. COMPSs workflows can be defined using Java, C++, or Python. COMPSs has been adapted to control the virtualization layer, making it transparent to the user, and also allowing to execute the same workflow in a series of environment, from single workstations, to HPC or grid/cloud facilities. Applications where the use of COMPSs would not be desirable can be also executed in their native environment, exploiting already existing parallelism if any.
Cloud applications
Not only dynamic virtual machines are supported, but also static web based applications, which can take profit of the HPC facilites if they need to perform heavy back-end calculations.
Cloud access
Access to MuGVRE local infrastructures is made accessible through the Programing Model Enacting Service (PMES). The user can interact with the service programmatically, and also via specifically designed web-based tools.
Web based access
DASHBOARD:
The web based tool offers a user-friendly interface that allows a full control of the infrastructure, and it is useful for managing small analysis and workflows. To obtain an account for the dashboard, please contact us.
guest : gu3st2014
DATA MANAGER:
The web utility allows to transfer I/O data to the MuGVRE cloud via HTTPS. If large transferences are required, accession through alternative vias are available. To consult them, or gain access to the application, please, contact us.

