Venue: Barcelona Supercomputing Center
Severo Ochoa room – Torre Girona building ( View in Google Maps )
Dates: 13th -14th December 2017
Participation: First-come, first-serve
Contact: Anna Montras
Registration Fee: 90€ . Included in the registration fee are coffee/snacks/refreshments and lunch served during workshop breaks. Participants are expected to cover their own travel and accommodation expenses.
Overview
This workshop will introduce delegates to a Virtual Research Environment (VRE), created by the Multi-scale complex Genomics project (MuG), to facilitate the analysis and interpretation of the 3D and 4D structure of the genome.
The MuG VRE integrates a range of data from genome annotation to 3D folding and DNA flexibility. Recent studies have shown the role that genome organisation can play in gene expression and the VRE has been designed to provide a way of analysing such data.
The course will explore how to use the multi-scale features and integrated tools of the MuG VRE through the study of real biological examples
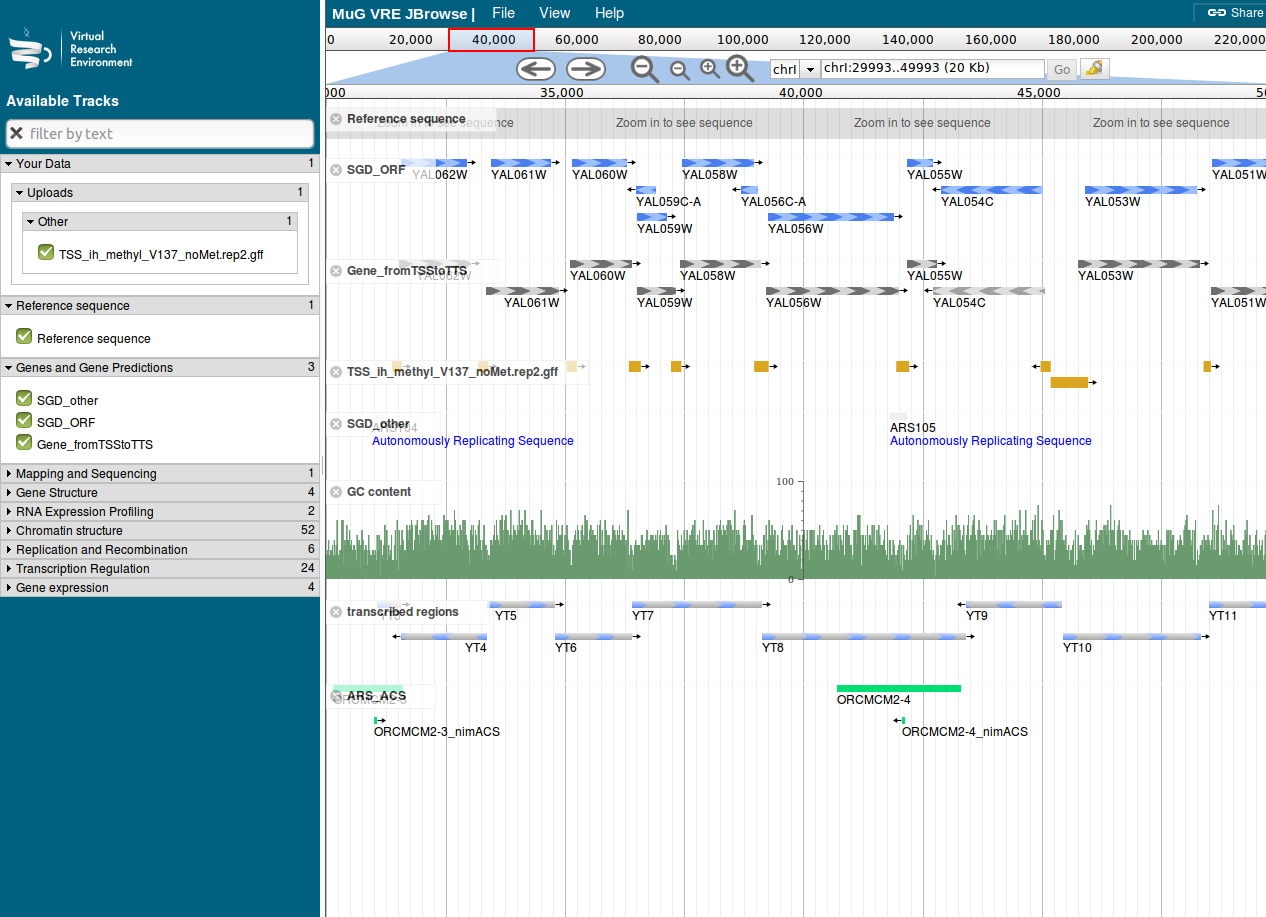
Figure 1: Nucleosome Dynamics
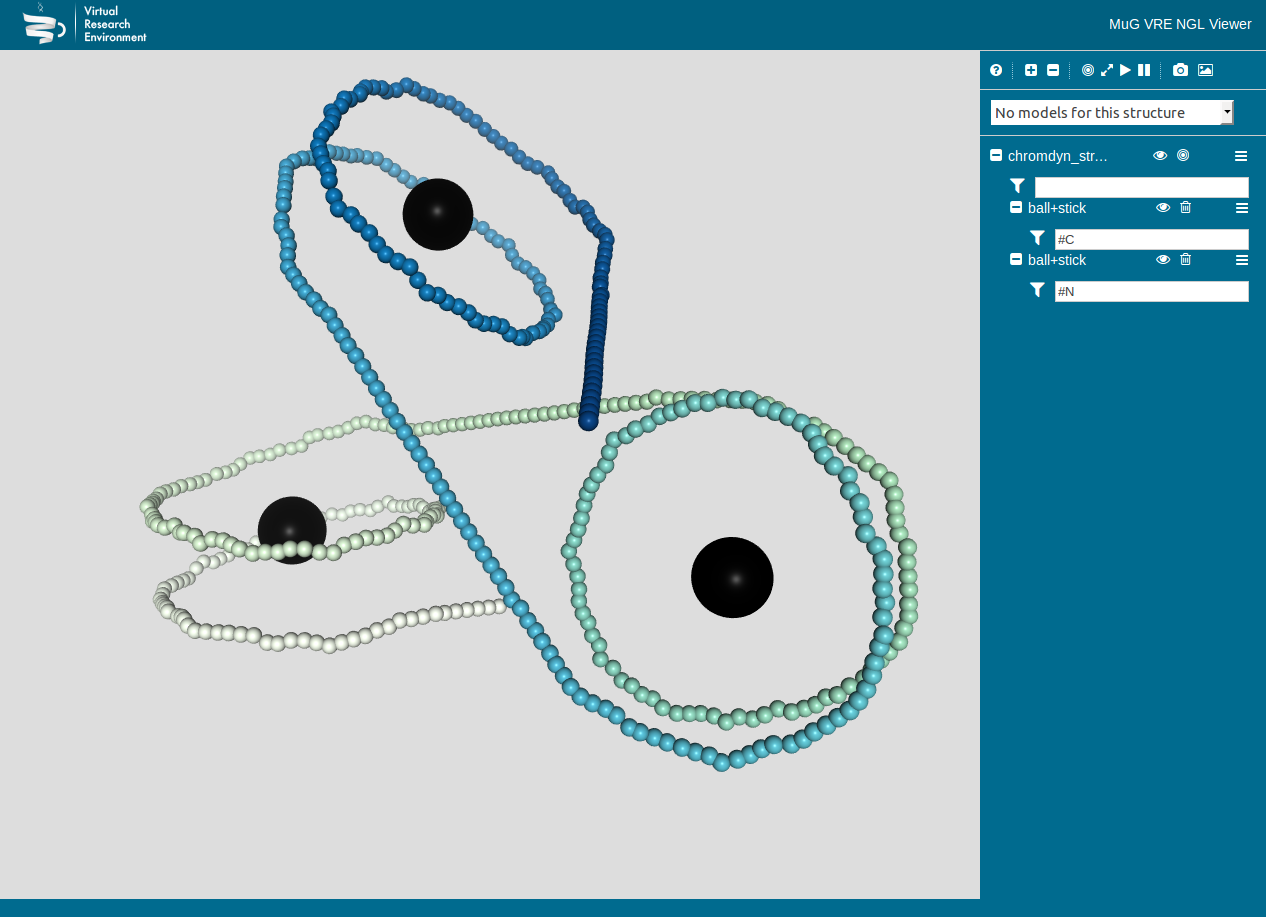
Figure 2: Chromatin Dynamics
Audience
The workshop is aimed at both experimental (bench-based) researchers and bioinformaticians; additionally it may be of interest to tool developers who wish to integrate their tools into the VRE.
Participants should have a Master’s degree or higher understanding of biology and have knowledge of genome organisation and the role it can play within the nucleus.
Requirements
Participants are expected to BRING THEIR OWN LAPTOP. (Please note an up-to-date version of Google Chrome is required to guarantee the correct performance of all tools featured in the course).
Syllabus, tools and resources
During this workshop participants will learn about:
- The Multi-scale complex Genomics project (MuG)
- The MuG Virtual Research Environment (VRE)
- Nucleosome dynamics
- Hi-C, interaction matrices and TADs (topologically associating domains)
- Protein-DNA interactions
Outcomes
After the workshop, participants will be able to:
- Navigate the MuG VRE
- Model and analyse a protein-DNA complex at the atomistic scale
- Process Hi-C data sets to produce interaction matrices and TADs
- Apply the MuG VRE to participants own data sets.
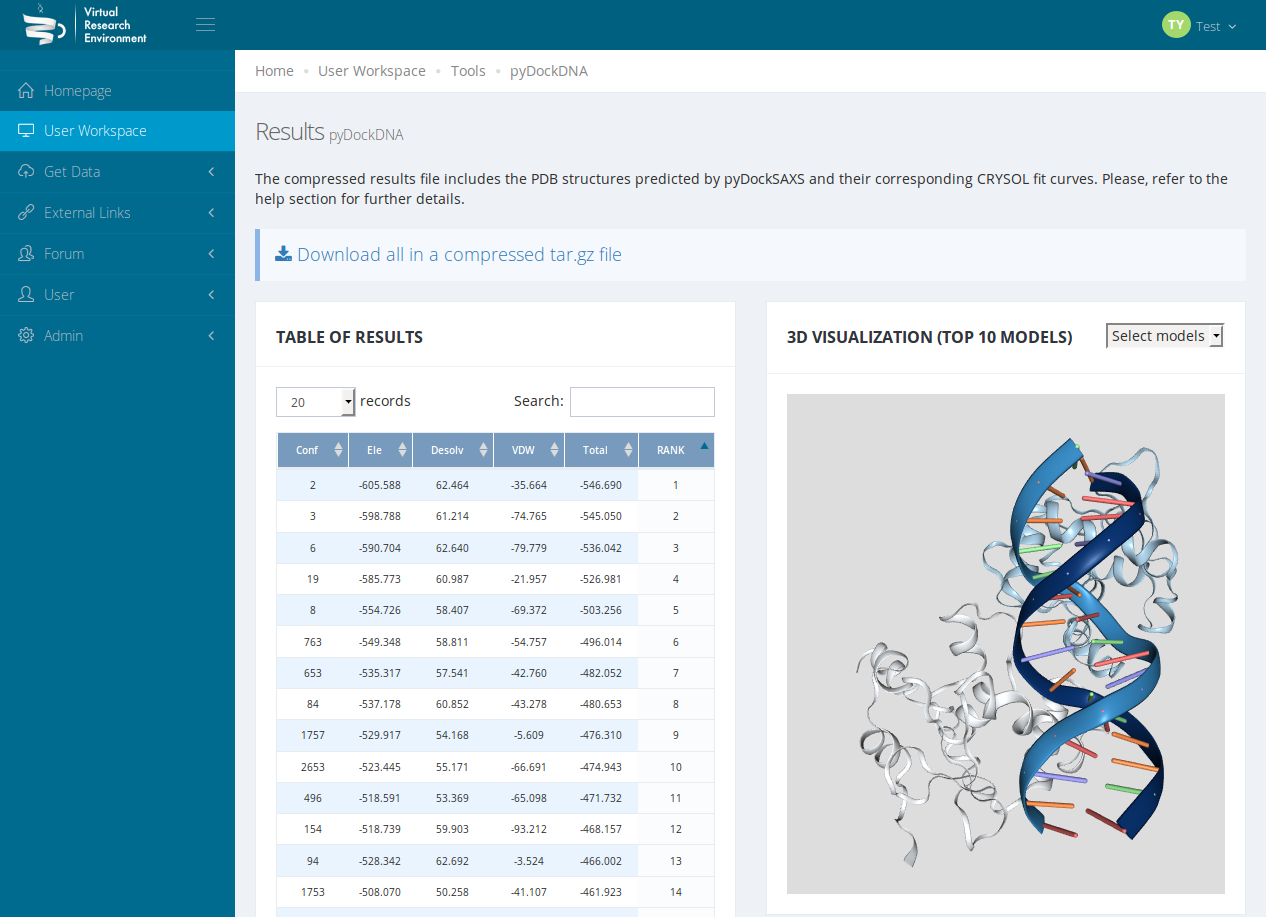
Figure 3: PyDock DNA
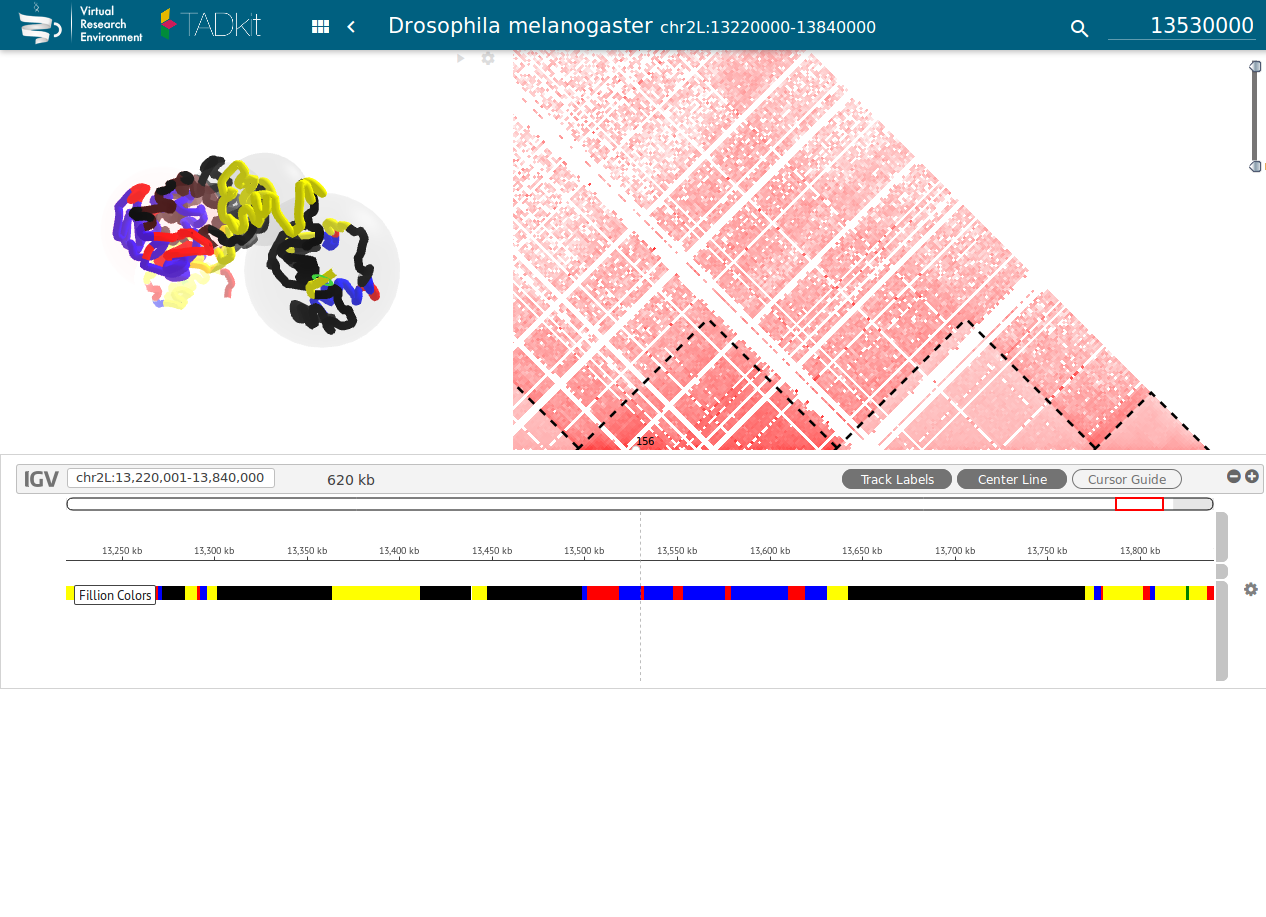
Figure 4: TADbit / TADkit
Programme
(*Preliminary programme subject to changes)
| Time | Topic | Instructor |
|---|---|---|
| Day 1 (Wednesday 13th December) | ||
| 9:00 – 9:30 | Registration and welcome | |
| 9:30 – 10:00 | MuG and participants introduction | Modesto Orozco |
| 10:00 – 10:30 | MuG VRE workspace presentation | |
| 10:30 – 11:30 | Presentation of Nucleosome Dynamics | Ricard Illa /Federica Battistini |
| 11:30 – 12:00 | Coffee Break | |
| 12:00 – 13:30 | Use case (application of Nucleosome Dynamics) | Ricard Illa / Federica Battistini |
| 13:30 – 14:30 | Lunch | |
| 14:30 – 16:00 | Coarse grained model of DNA. Build and analyze a sequence of interest. | Jürgen Walther |
| 16:00 – 16:30 | Coffee Break | |
| 16:30 – 17:30 | Coarse grained model of chromatin; Generation and analysis of chromatin fiber structures. | Jürgen Walther |
| 17:30 – 18:00 | Day 1 wrap-up , open questions and answers | |
| Day 2 (Thursday 14th December) | ||
| 9:00 – 10:45 | Presentation of Protein-DNA and protein-protein docking tools | Brian Jiménez |
| 10:45 – 11:15 | Coffee break | |
| 11:15 – 13:00 | Protein-DNA interactions, DNA flexibility and binding specificity. | Charles Laughton |
| 13:00 – 14:00 | Lunch | |
| 14:00 – 15:30 | Presentation of TADbit: generation of Hi-C interaction matrices from Hi-C data | David Castillo |
| 15:30 – 16:00 | Coffee break | |
| 16:00 – 17:30 | Generation of populations of structures that satisfy the Hi-C interaction matrices. Visualization of 3D models using TADkit | David Castillo |
|
17:30 – 18:00 |
Course wrap up and feedback | |
With the support from:


